Welcome to MethGET!
MethGET is a web-based software for analyzing the correlation between genome-wide DNA methylation and gene expression. Stand-alone version can also be available on https://github.com/Jason-Teng/MethGET.
References to cite MethGET:
Teng CS, Wu BH, Yen MR, Chen PY. MethGET: web-based bioinformatics software for correlating genome-wide DNA methylation and gene expression. BMC Genomics. 2020 May 29;21(1):375. doi: 10.1186/s12864-020-6722-x.
https://pubmed.ncbi.nlm.nih.gov/32471342/
Contact information:
Jason Teng (faf046059@gmail.com)
Features

Analyses Examples
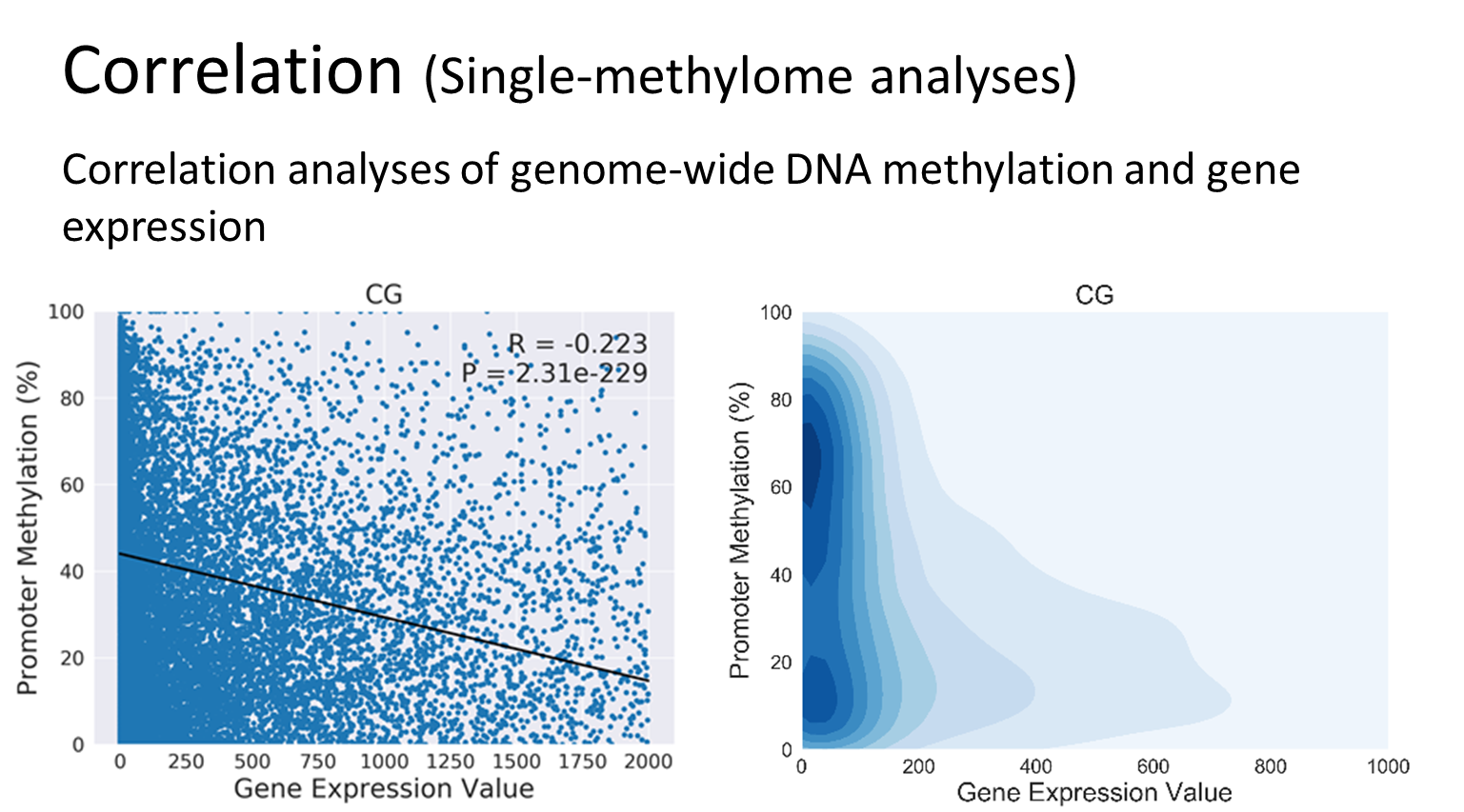
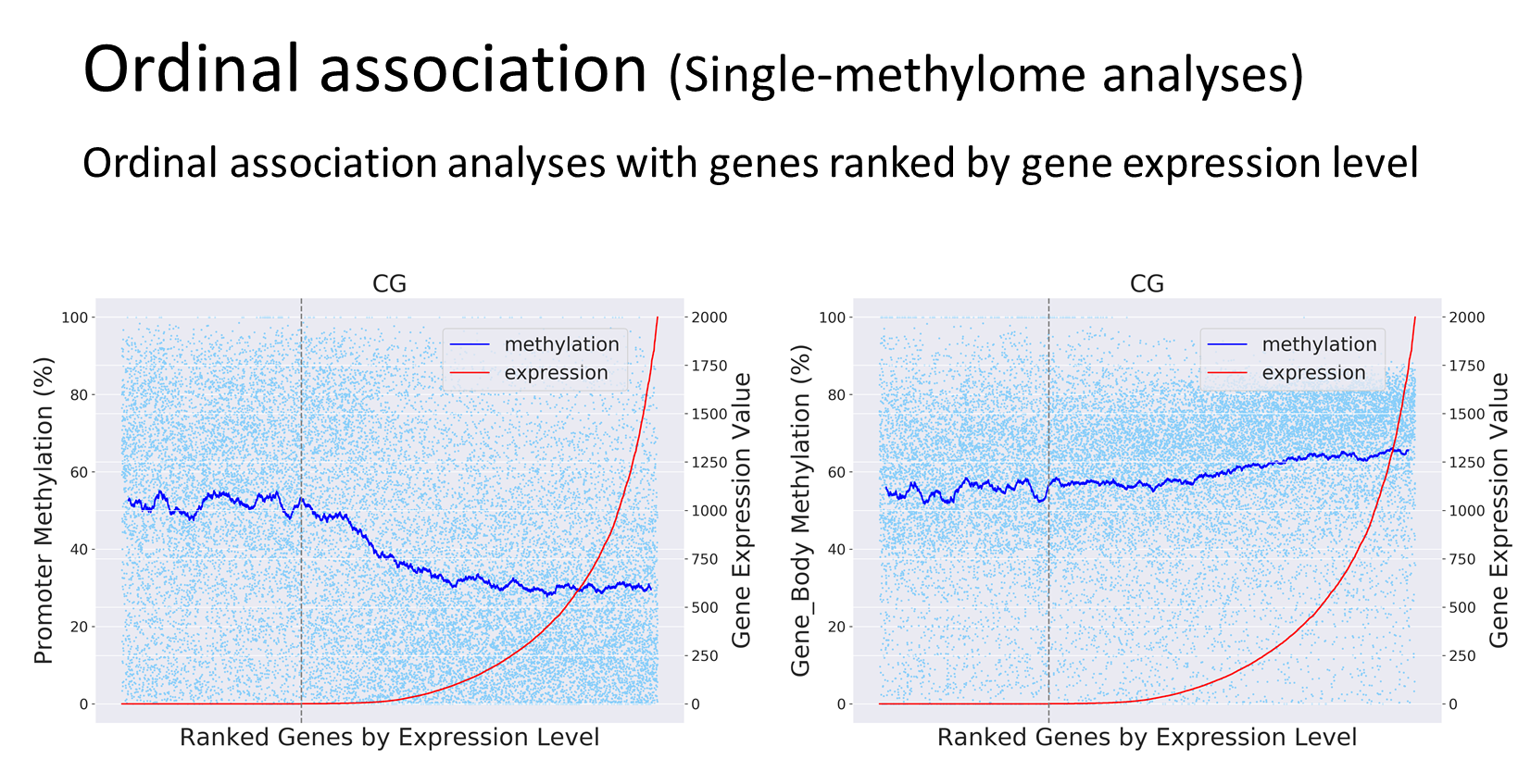
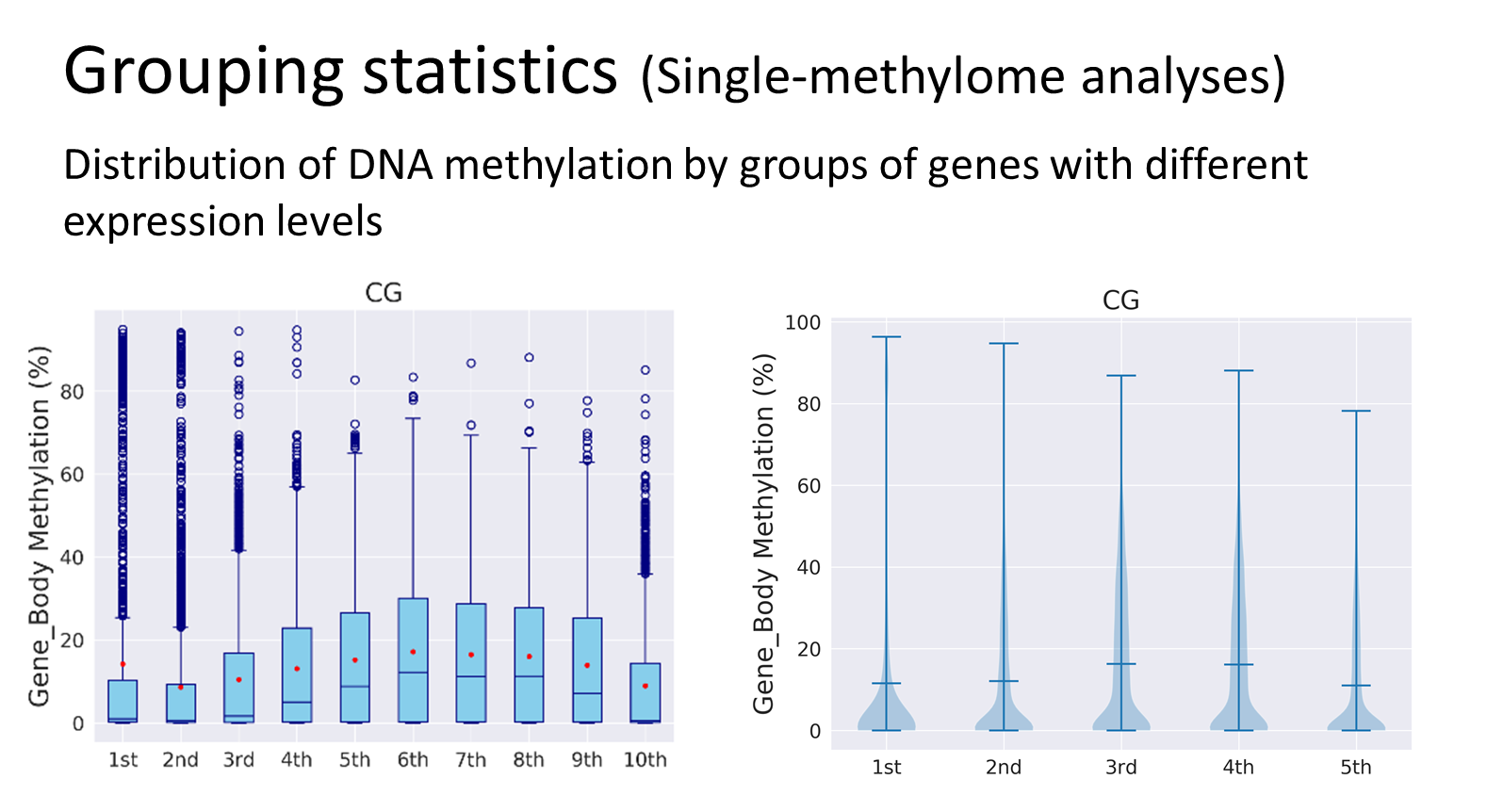
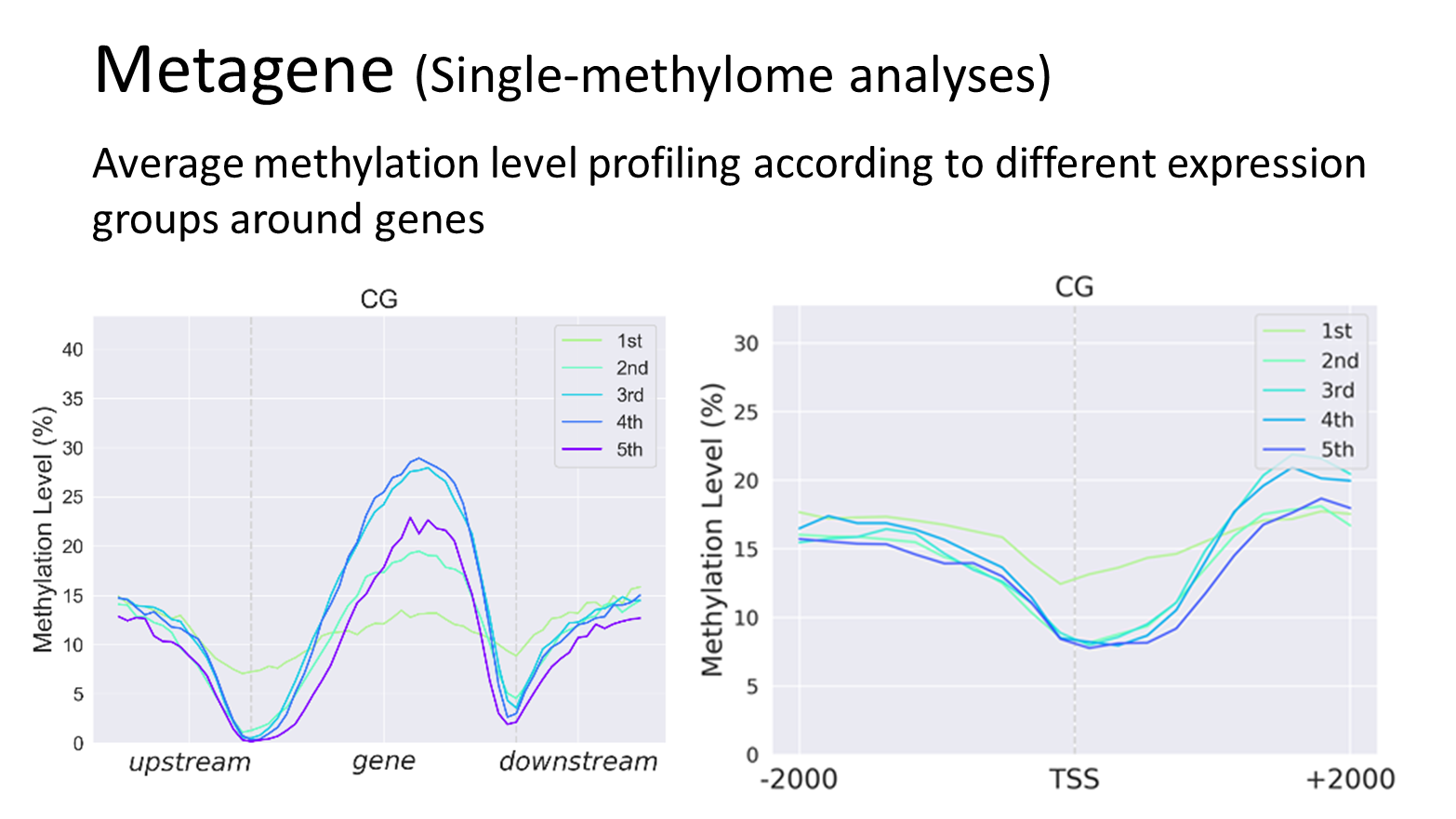
Quick Guide
1. Upload data
- Enter Your Project Name and Email.
- Upload DNA Methylation (CGmap.gz) and Gene Expression Files (Exp.txt).
- Choose The Organism.
- Click “submit” button.
*you will receive 2 emails.
(1) start preprocessing mail (2) finish preprocessing mail (get account name)2. Launch analysis
- sign in the account name (example account is provided).
- choose the parameters of analysis.
- submit (minutes).
3. GET results
- see results of CG, CHG, CHH.
- download selected figures and tables or just download all.
Note
Single methylome: check the checkbox of the sample and upload a CGmap.gz and a Exp.txt
(0.5hr per methylome for Arabidopsis)
metagene: check the checkbox of metagene (3.5hr per methylome for Arabidopsis)
multiple-methylome: need at least one methylome in both group A and group B
Input Format
DNA methylation data: CGmap
CGmap files are the output of BS-Seeker2 aligning pipeline that contain information of DNA methylation level, read counts and methylation context of each cytosine. The instruction for generating CGmap can be freely available from here. The methylation calling files from other aligners/callers, MethGET provides a python script (methcalls2CGmap.py) to convert them to CGmap.gz.
| Chr1 | G | 125 | CHG | CA | 0.82 | 9 | 11 |
| Chr1 | G | 126 | CHH | CC | 0.18 | 2 | 11 |
| Chr1 | G | 129 | CHH | CA | 0.33 | 3 | 9 |
| Chr1 | C | 135 | CHH | CA | 0 | 0 | 12 |
| Chr1 | C | 161 | CG | CG | 0.76 | 16 | 21 |
| ... | ... | ... | ... | ... | ... | ... | ... |
Gene expression data: Tab-delimited text file
Tab-delimited text files contain the gene names and gene expression values (with no header in the first line). Official gene symbol is suggested to fit our build-in gene annotation.
| AT1G01610 | 34.2495 |
| AT1G01620 | 453.344 |
| AT1G01630 | 53.2985 |
| AT1G01640 | 40.7508 |
| AT1G01650 | 36.3414 |
| ... | ... |
Gene annotation: GTF file
GTF files contain gene names and transcript annotation of the genome that can be available from ensemble FTP. The gene annotations for Arabidopsis, Rice, Human, and Mouse have built on the web.